Welcome to the live instance of the European Galaxy server. This is your gateway to start interactive tools, such as Jupyter, RStudio or other web-applications. Each Interactive Tool runs in its own container on the de.NBI-cloud. Every registered user can start up to 10 different interactive tools simultaneously and can keep them running for up to 30 days. You can interact with Galaxy via the API and in Jupyter and RStudio we have added convenient functions (put/get) to transfer data back and forth between Galaxy and Jupyter resp. RStudio. Of course, you can also store your entire Notebook or R session back to Galaxy and save all provenance information in a permanent storage.
If you start an Interactive tool it will keep spinning in a yellow state as long as the Container is running. To open your Tool you open the link at User → Active InteractiveTools. The eye icon of your Galaxy dataset is not working yet. A Jupyter and RStudio tutorial is provided by the Galaxy training Network.
Enjoy!

Jupyter lab for Python, R and Julia

RStudio with basic R packages

Jupyter Notebooks for the MGnify API

SPARQL query interface
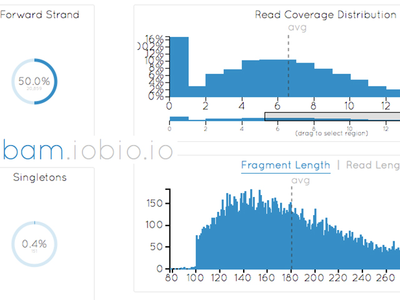
BAM IOBIO
Examine sequence alignment in seconds
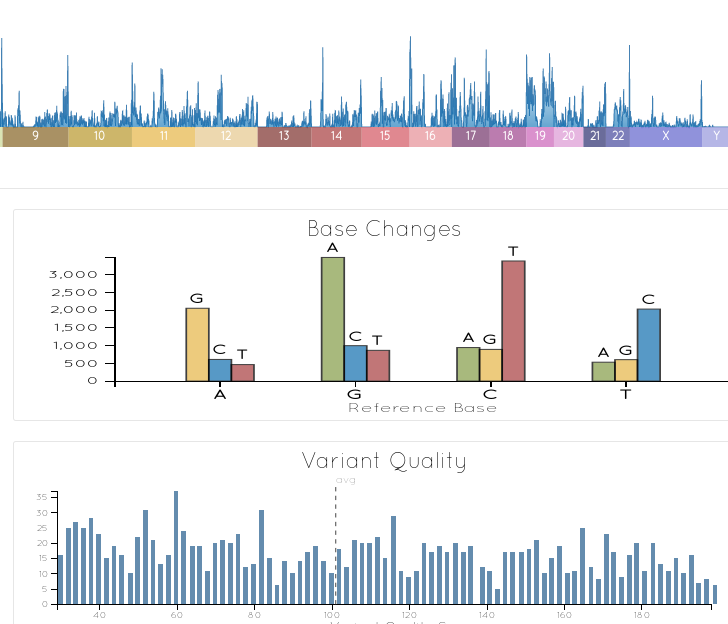
VCF IOBIO
Examine your variant file in seconds

Explore single-cell transcriptomics data
Ethercalc
A web spreadsheet
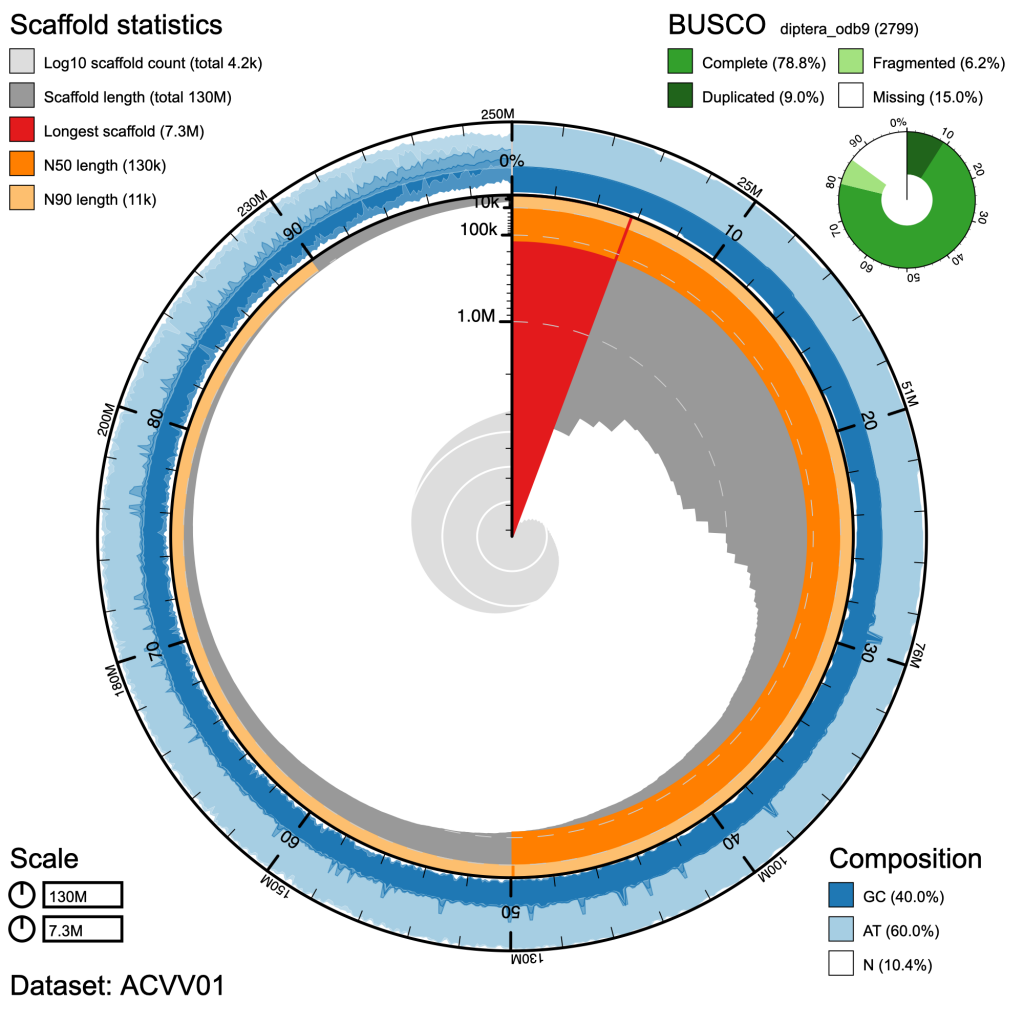
BlobToolKit
Genome-scale dataset visualistion

Visualise large biological datasets (microbiomes, metagenomes, etc.)

MetaShARK
Generate Metadata from data using EML standard
Wallace
Model niches and distributions of species

iSEE - SummarizedExperiment Explorer

Webbased Interactive Omics visualization
IDE for Computational Materials Science
Virtual Desktop environment

Jupyter lab for ocean / atmosphere / land / climate python ecosystem

Panoply netCDF Data Viewer

HiGlass