usegalaxy.eu
Our Galaxy server (https://usegalaxy.eu) is the biggest Galaxy instance in Europe, and one of the biggest worldwide.
With this server, we provide access to:
- a huge compute and storage resource free of charge,
- more than 3900 different, well-documented and constantly maintained scientific tools,
- 250+ GB per user (500 GB for ELIXIR members).
- We also support numerous additional storage options and you can learn about them here.
When this effort is combined with our community-maintained workflows and our in-depth training material, it makes up for a truly productive work experience. We believe in enabling everyone to perform reproducible science.
ACKNOWLEDGEMENT
We are aiming to maintain high competency and provide high-quality data analysis services to all our Galaxy users.
Therefore, we request that you acknowledge this service by including the members of the Freiburg Galaxy Team as co-authors if they have made a significant intellectual and/or organizational contribution to the work described (conceptualization, design, data analysis, data interpretation and/or input into drafting, revising or writing any portion of the manuscript).
Individuals who have contributed to the project, but whose contributions do not rise to the level justifying authorship, can be recognized in the acknowledgements section of the manuscript as follows:
The authors acknowledge the support of the Freiburg Galaxy Team: Person X and Björn Grüning, Bioinformatics, University of Freiburg (Germany), funded by the German Federal Ministry of Education and Research BMFTR grant 031 A538A de.NBI-RBC and the Ministry of Science, Research and the Arts Baden-Württemberg (MWK) within the framework of LIBIS/de.NBI Freiburg.
Additional funding of projects as well as the provision of material expenses, is welcome to support our growing Galaxy community in Freiburg and Europe. Please also cite the main Galaxy publication.
Data Analysis for any Scientist
No programming knowledge is required to access Galaxy and its tools; all you need is a web browser. It provides robust data analysis and can replace Excel habits.
The computation is automatically performed on a cluster and on the de.NBI cloud. Each user can store 250 GB (500 GB for ELIXIR members) for analyses (and even more on request).
For advanced users, https://usegalaxy.eu can be programmatically accessed via the API to leverage its cluster and data storage to scale beyond the visual interface and to automate the analyses. Novel analyses can also be developed right in the interface with Galaxy Interactive Environments: Jupyter notebooks, Ethercalc, HiCBrowser, …
Visualization & Data Interpretation
Publication-ready visualizations can be instantaneously generated inside Galaxy: charts with bar diagrams, line charts, box plots, heatmaps, scatter plots, Venn diagrams, and much more.
To dig deeper, interactive visualizations are available:
- Integrated Genome Browser: Trackster
- Phinch for BIOM files, Circos, JBrowser, IGV, …
Tools for any Bioinformatics Analysis
More than 2500 scientific tools can be accessed via https://usegalaxy.eu, covering most of the bioinformatics topics:
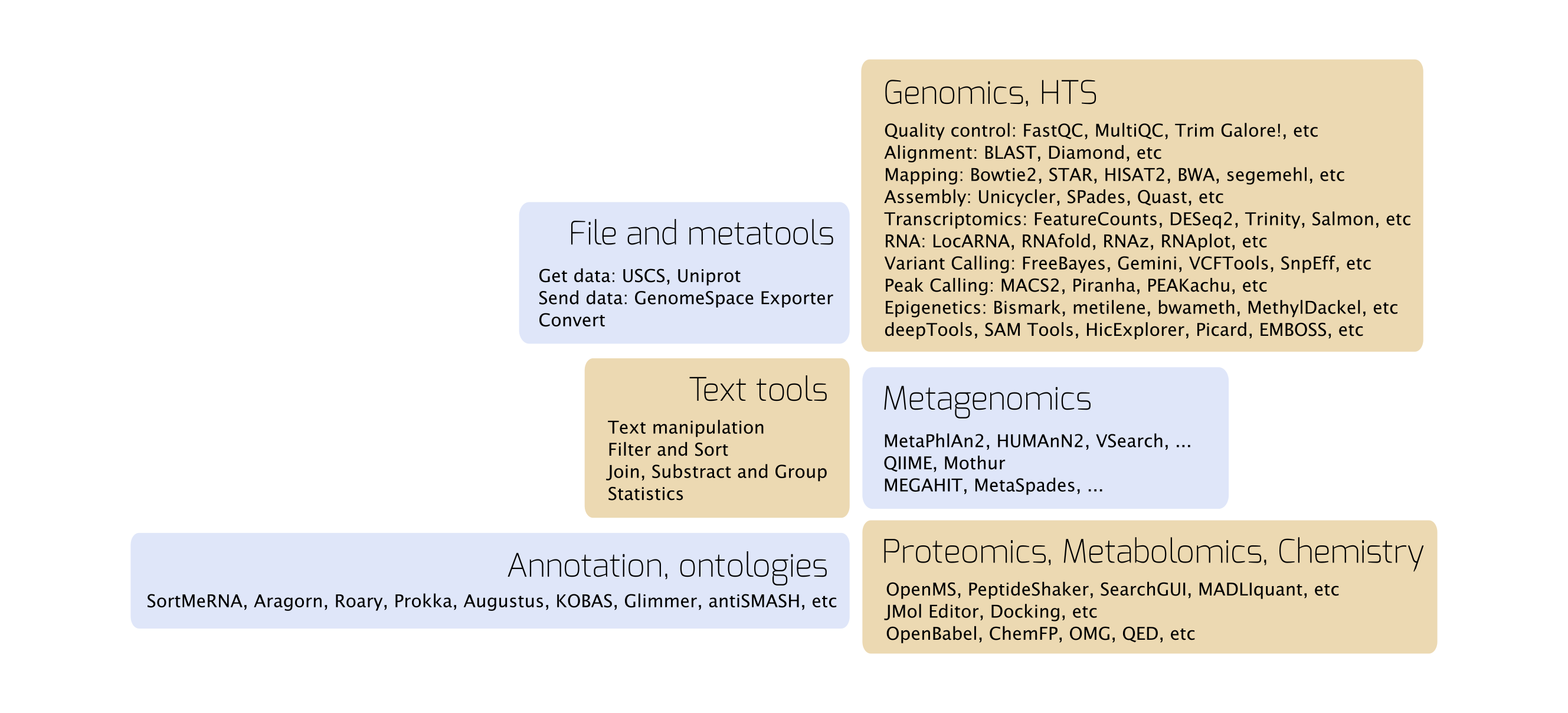
The tools and analysis pipelines are regularly updated.
Reproducibility & Transparency
The history is the foundation of reproducibility and transparency in Galaxy. It captures inputs, parameters, and versions of the used tools. It can be shared with everyone, even outside the Galaxy framework.
Galaxy also provides a powerful workflow system. Workflows can be created by extracting workflows from histories or from scratch with drag-and-drop. They are downloadable and shareable with everyone, with no vendor lock-in.
Thousands of tools are available with fixed versions and are managed by Bioconda and BioContainers.
4 TB of reference data are available on https://usegalaxy.eu providing access to hundreds of reference genomes.
Supported communities with their own subdomain
- ecology.usegalaxy.eu
- hicexplorer.usegalaxy.eu
- metagenomics.usegalaxy.eu
- rna.usegalaxy.eu
- proteomics.usegalaxy.eu
- clipseq.usegalaxy.eu
- streetscience.usegalaxy.eu
- graphclust.usegalaxy.eu
- cheminformatics.usegalaxy.eu
- imaging.usegalaxy.eu
- singlecell.usegalaxy.eu
- climate.usegalaxy.eu
- nanopore.usegalaxy.eu
- metabolomics.usegalaxy.eu
- humancellatlas.usegalaxy.eu
- annotation.usegalaxy.eu
- erasmusmc.usegalaxy.eu
- live.usegalaxy.eu
- plants.usegalaxy.eu
- ml.usegalaxy.eu
- virology.usegalaxy.eu
- covid19.usegalaxy.eu
- africa.usegalaxy.eu
- microbiome.usegalaxy.eu
- cancer.usegalaxy.eu
