New generation of sequencing platforms coupled to numerous bioinformatics tools has led to rapid technological progress in metagenomics and metatranscriptomics to investigate complex microorganism communities. Nevertheless, a combination of different bioinformatic tools remains necessary to draw conclusions out of microbiota studies. Modular and user-friendly tools would greatly improve such studies.
We therefore developed ASaiM, an Open-Source framework dedicated to microbiota data analyses. ASaiM integrates a comprehensive set of microbiota related tools, predefined and tested workflows dedicated to microbiota analyses. ASaiM offers then sophisticated analyses to scientists without command-line knowledge and a powerful framework to easily and quickly explore microbiota data in a reproducible and transparent environment.
ASaiM is also available as a Docker Galaxy flavour, but tools and workflows can be tested here.
Read more on ASaiM’s homepage or via its preprint
Workflows
Several workflows has been developed for ASaiM and are available on
-
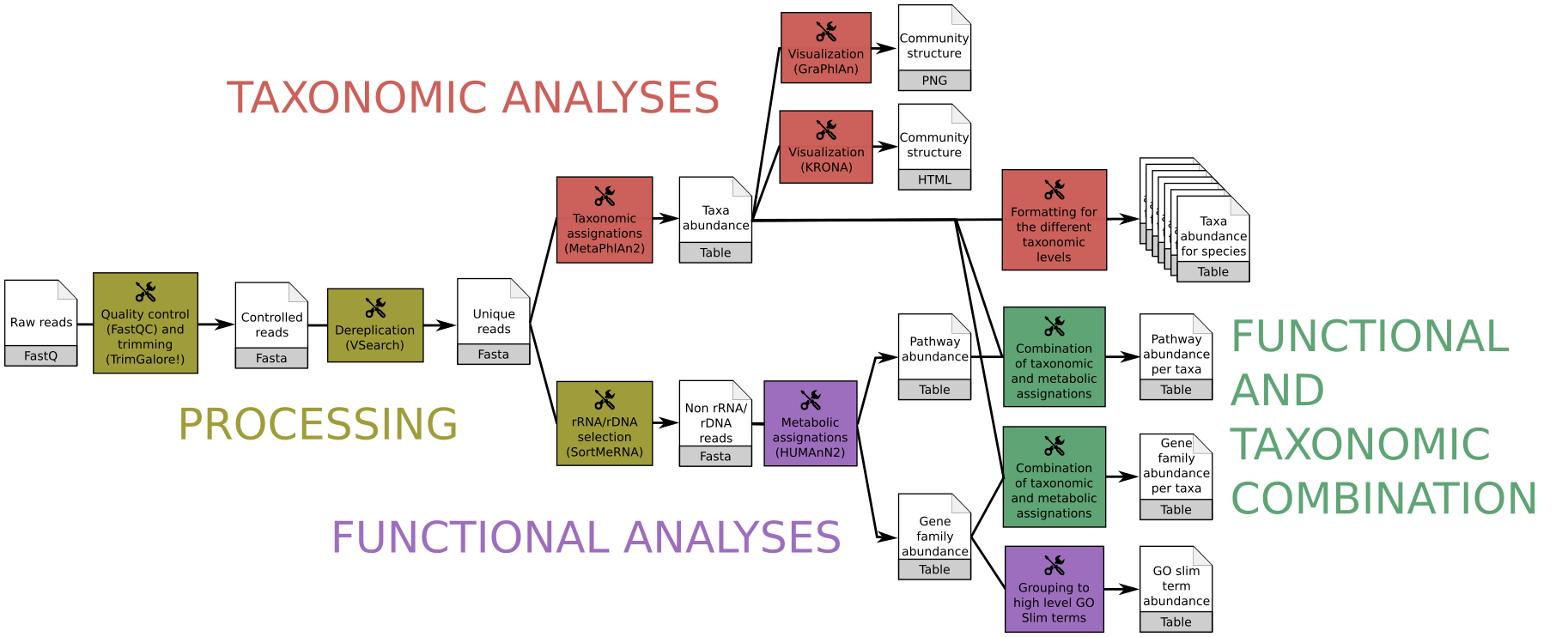
Analysis of raw metagenomic or metatranscriptomic shotgun data: The workflow quickly produces, from raw metagenomic or metatranscriptomic shotgun data, accurate and precise taxonomic assignations, wide extended functional results and taxonomically related metabolism information
- Assembly of metagenomic data with two workflows, each one using one of each of the well-performing assemblers: MEGAHIT and MetaSPAdes
- Analysis of metataxonomic data with QIIME - Illumina Overview tutorial
- Running as in EBI metagenomics (3.0) to reproduce locally analyses made in EBI Metagenomics website, without having to wait for availability of EBI Metagenomics or to upload any data on EBI Metagenomics
