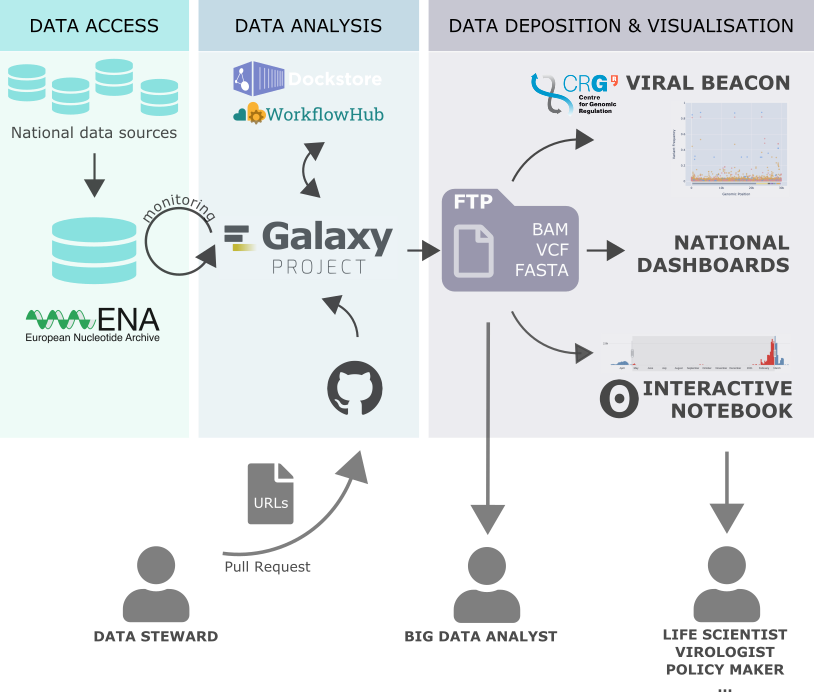
Recently, we started a large-scale project on the European Galaxy server for automatically collecting, and analyzing raw sequencing data published by the COVID-19 Genomics UK Consortium (COG-UK) and, potentially, other large-scale SARS-CoV-2 sequencing projects. As a unique feature, we aim to make all analysis steps leading from sequenced reads to lists of genomic variants and consensus sequences fully open and accessible.
Automated variant calling workflows
Every three hours, newly published COG-UK samples are processed to produce:
- public Galaxy histories, which provide full data provenance
- variants in VCF format
- alignment files in BAM format
- consensus sequences in FASTA format
These are subsequently pushed back to a public FTP server hosted by CRG/BSC. CRG then takes this data and integrates it into the Viral Beacon.
The scripts for the workflow automation are publicly available together with documentation. The analysis is based on public, validated workflows that are available via workflowhub.eu.
Special care was taken to run this analysis on scale without disrupting normal Galaxy usage. In other words, we can monitor samples, and at the same time still run trainings and serve hundreds of other Galaxy users simultaneously.
If you want us to process samples from your country, too, please get in touch with us and we will help you get things running!
Interactive ObservableHQ dashboard
Furthermore, all data that we process is summarized in an ObservableHQ dashboard. Anyone can access, play around with the data in JavaScript and JSON and push any new plots they create back to us.
Since it’s a notebook, you can also filter the data with real-time queries. At the lower end, under “variants of concern”, you can for example select your favorite lineage and see how often its signature mutations are identified. As in most countries, the South African and the Brazilian variants aren’t really taking off in the UK. The Bloom DMS Escape variants also aren’t seen much. These are variants that have been demonstrated in lab experiments to cause immune escape. So good news for the UK - what about the other European countries? Well, for this we need your Galaxy instances to analyze the other data - and maybe more importantly, we need more OPEN DATA!
Request processing of your samples of interest
In addition, we wanted to make this automated resource useful for everyone, including non-Galaxy users. To this end, we have opened up an on-demand analysis service via a simple GitHub repository: https://github.com/usegalaxy-eu/sars-cov-2-processing-requests. The idea is very simple; you just submit a file with URLs (ENA URLs for example) as a single file. This file makes up one batch of processing requests.
Members of the European Galaxy Team will check your contribution and approve it, after which Galaxy will pick those files up and process them with our standard pipelines.
Just as for the automated COG-UK data analysis, all files are pushed back to the CRG FTP server. So in the end, after a few hours (depending on the amount of URLs and the load on our servers), you can access the results through either:
- Galaxy via shared/public histories
- via the CRG FTP server
and see them visualized on the Viral Beacon dashboard (with a delay of probably a few days).
This should make it even easier to analyze viral sequences with Galaxy, in particular, for people who don’t want to run their own workflows, but are still interested in specific batches of samples not covered by our automated monitoring efforts.


