The BM425 Advanced Microbiology module at the University of Strathclyde is a final-year course that aims to help students understand the genomic basis, molecular biology, structure and function of microbial systems and their interactions with the environment.
The first block of the course covers genome sequencing, assembly, and comparative genomics, in an environmental and clinical context. In the first week, the students participate in a workshop where they take a set of raw reads through quality control, assembly and annotation, and contextualise their organism using comparative genomics. We teach this using Galaxy as many of the cohort are unfamiliar with the command-line, and we need a consistent, common interface that allows them to do real science, while minimising the intimidation and complexity of using multiple different tools.
Like everyone else, for the 2020 course presentation we were challenged by the global coronavirus pandemic. In particular, we could not run our usual face-to-face Galaxy workshop in the on-campus computer labs. We were also looking to move on in some way from our on-site teaching hardware, which struggled under the load of an entire class assembling their reads in the previous year’s presentation. We aim to be relevant and timely in our teaching so, for this year’s course, we were asking the students to assemble and annotate the Wuhan SARS-CoV-2 isolate, and compare it with a 2003 SARS isolate, and a more recent Dutch SARS-CoV-2 isolate.
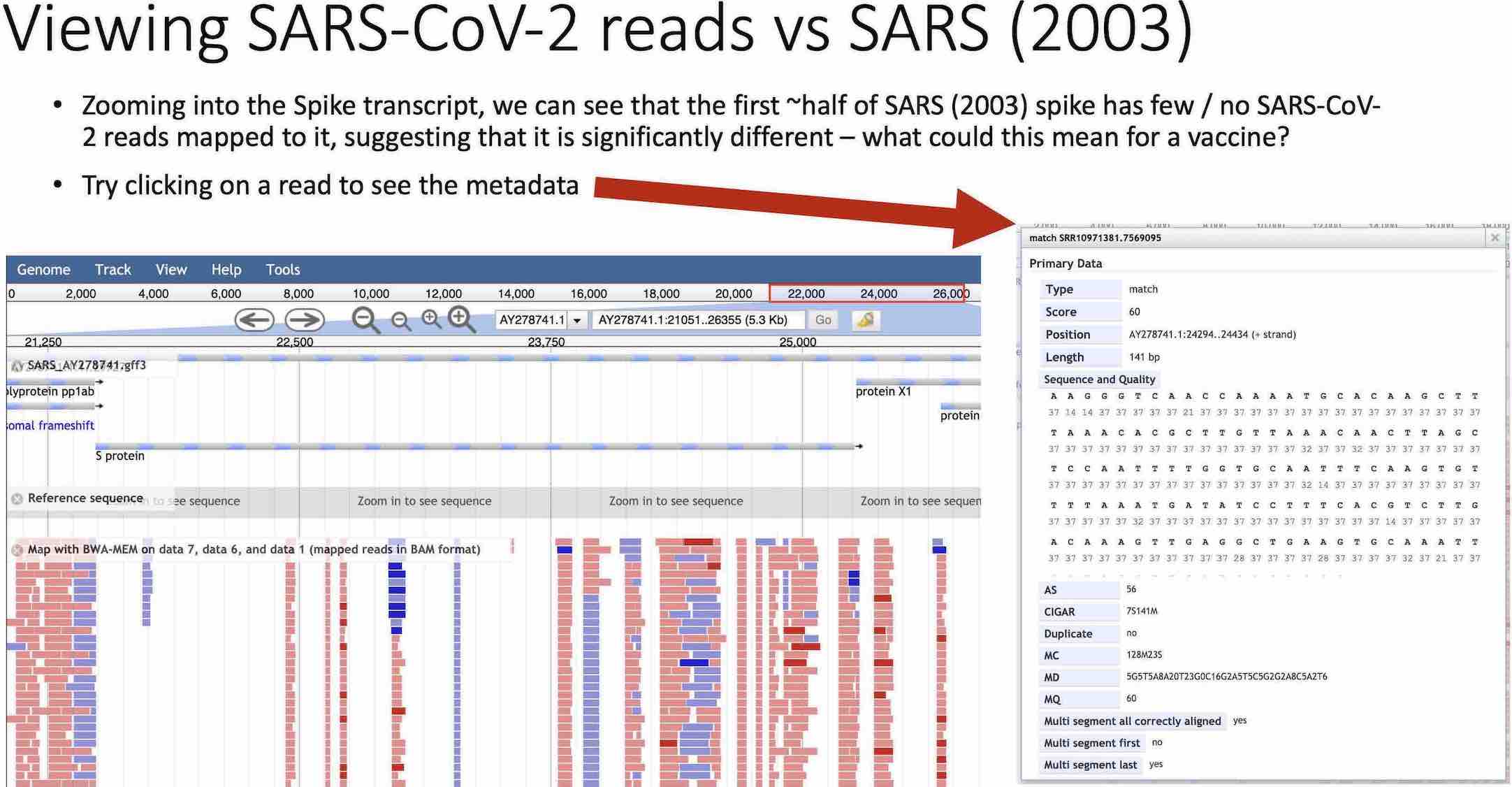 Slide from the workshop guide, comparing SARS-CoV-2 mapping to a SARS genome
Slide from the workshop guide, comparing SARS-CoV-2 mapping to a SARS genome
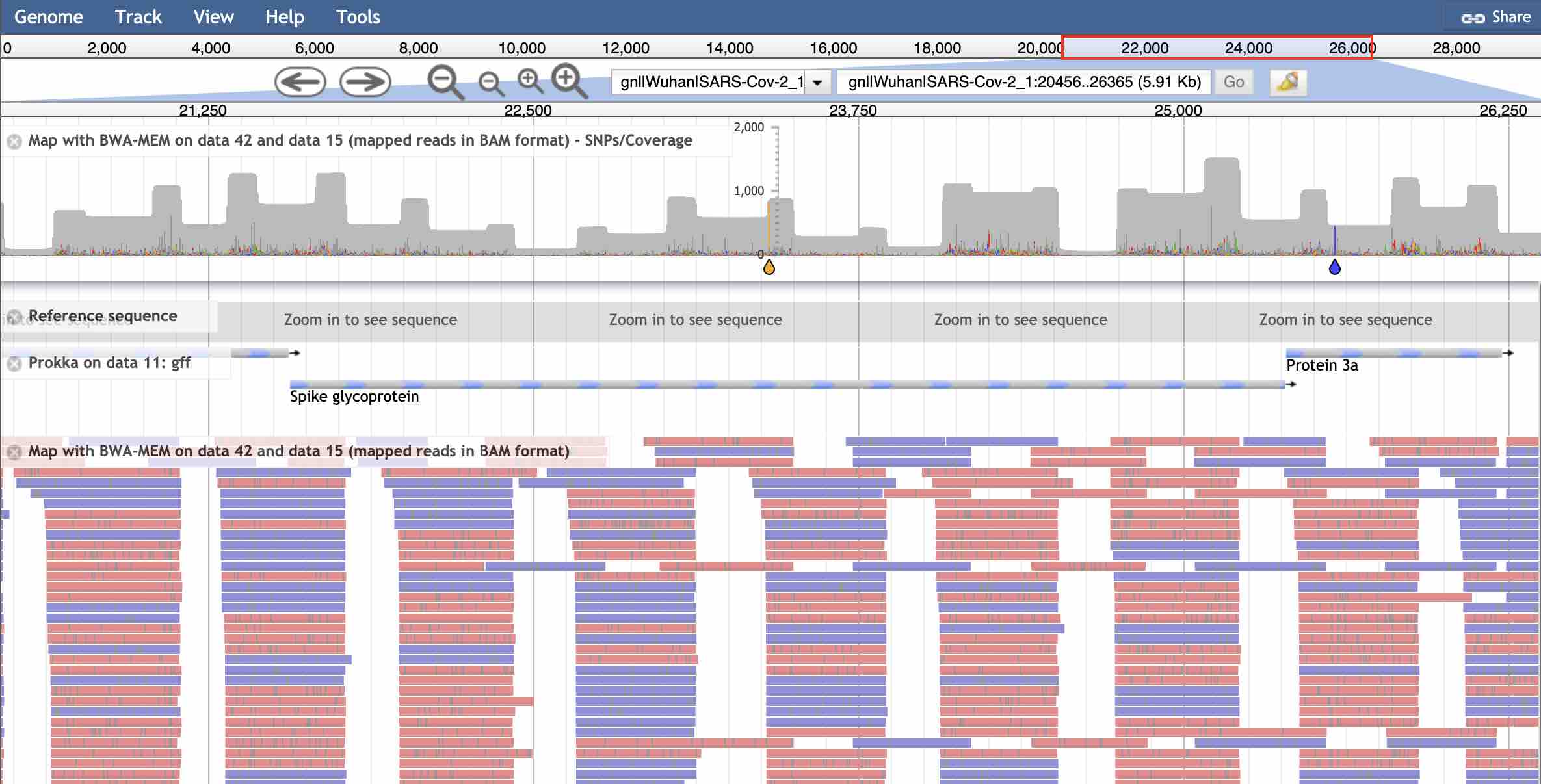 JBrowse view of Dutch nanopore reads mapped to the Prokka-annotated assembly of the Wuhan SARS-CoV-2 genome. Coverage chart & variants at the top – note the characteristic pattern caused by overlaps of the amplicons and the SNP in the Spike. Annotation track is the GFF file from Prokka, bottom window shows the individual reads.
JBrowse view of Dutch nanopore reads mapped to the Prokka-annotated assembly of the Wuhan SARS-CoV-2 genome. Coverage chart & variants at the top – note the characteristic pattern caused by overlaps of the amplicons and the SNP in the Spike. Annotation track is the GFF file from Prokka, bottom window shows the individual reads.
We needed to find a way to deliver this course so that ≈50 geographically-separated students and course tutors could simultaneously work in a stable environment with minimal queuing and hardware issues, so that we could focus on teaching, rather than administration of the computing resource. We also needed to account for a certain amount of asynchronous participation, so that we could test the materials, and allow for students with other time pressures due to lockdown to progress through the material in their own time.
We became aware of the Freiburg Galaxy TIaaS service over the Summer break, and it looked to be perfect for our use case. The application process was straightforward and Beatriz was extremely helpful in guiding us through the process; the gentle nudge regarding equivalent cost of AWS provision is also useful information for planning future teaching budgets.
TIaaS generously assigned to us 9 dedicated servers for the duration of the course, each with 32 cores and 240GB of RAM. All students and tutors were able to register simply for the private queue by visiting the course link; all other interactions were transparent and identical to day-to-day https://usegalaxy.eu/ usage.
On the day, the service ran smoothly for ≈44 simultaneous students and staff, presenting no hardware or queuing problems. The Galaxy service purely facilitated teaching and did not place any major computing hurdles in the way of student progress. As tutors, we also had access to a queue status page through which we could watch the jobs as they ran. This was very useful as we could get a sense of the class’s progress. We could also anticipate questions and issues (which we handled in Zoom breakout rooms, sharing screens where necessary), because the failing jobs were visible. More curious students could also be directed to the Galaxy Training Materials site, having gained some confidence in the interface.
The student feedback was overwhelmingly positive. Many of the students had not had such detailed exposure to bioinformatics of NGS sequencing, and we had specific feedback reporting that the Galaxy environment and materials took away the fear and anxiety of working with an unfamiliar and, to some, daunting topic. The smooth running of TIaaS meant that students and staff could focus entirely on the science and teaching, which was a tremendous help, and we are extremely grateful for the support provided by Galaxy Europe and the Freiburg Galaxy Team.
Leighton Pritchard, Nick Tucker, Morgan Feeney, Paul Herron (BM425 Bioinformatics Workshop)
[1] Wu, F., Zhao, S., Bin Yu, Chen, Y.-M., Wang, W., Song, Z.-G., et al. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. http://doi.org/10.1038/s41586-020-2008-3
[2] Munnink, B. B. O., Nieuwenhuijse, D. F., Stein, M., O’Toole, Á., Haverkate, M., Mollers, M., et al. (2020). Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nature Medicine, 382, 1–6. http://doi.org/10.1038/s41591-020-0997-y

