We have received some feedback about TIaaS from Abdus Salam:
A week in Genomics
Identification of Single Nucleotide Polymorphism from sequencing reads by using Galaxy server.
Speaker: Abdus Salam
Held on: 26-30 April 2021
Venue: Precision Medicine Lab, Rehman Medical Institute, Hayatabad, Peshawar, Pakistan.
To begin with, I would like to show my gratitude on behalf of all the members who attended the training session and the organizing team especially Saad Zaheer who made the overall process smooth and fun to attend. Researcher and students from diverse backgrounds of Engineering, Biotechnology, Computer Science, Medicine, and Biology attended our training session and lauded and showed their gratitude to the Galaxy team for their generous support of providing us with their on-cloud computational resources. The session offered a thorough analysis of Head and Neck Squamous Cell Carcinoma sequencing read files and variant identification in comparison with reference files. For those who have no prior experience of the domain of Computational Biology, workflow design and analysis of Whole Exome Sequencing data is a job that requires a lot of computational tools and resources and effort, knowledge and caution on user’s behalf. Galaxy made that extremely effortless for us by offering their classy tools, on-cloud storage, memory, and processing power, and an easy to use and fun interface. The coolest bit about Galaxy is that it allows you to view output files generated after performing analysis on input files. Following are a few snapshots of the analysis we performed.
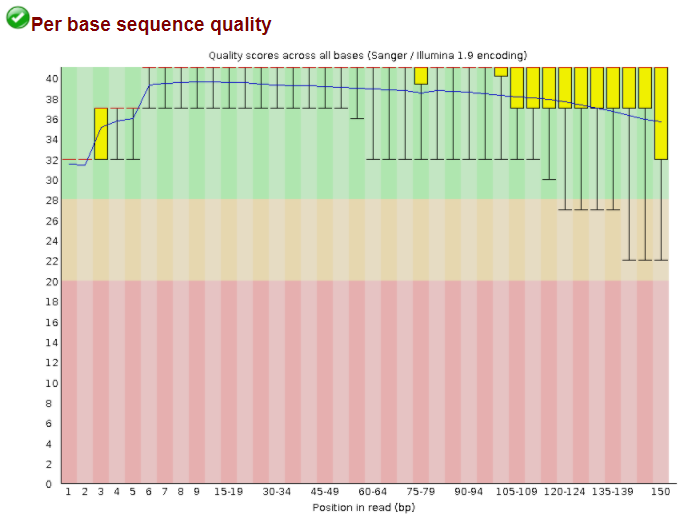
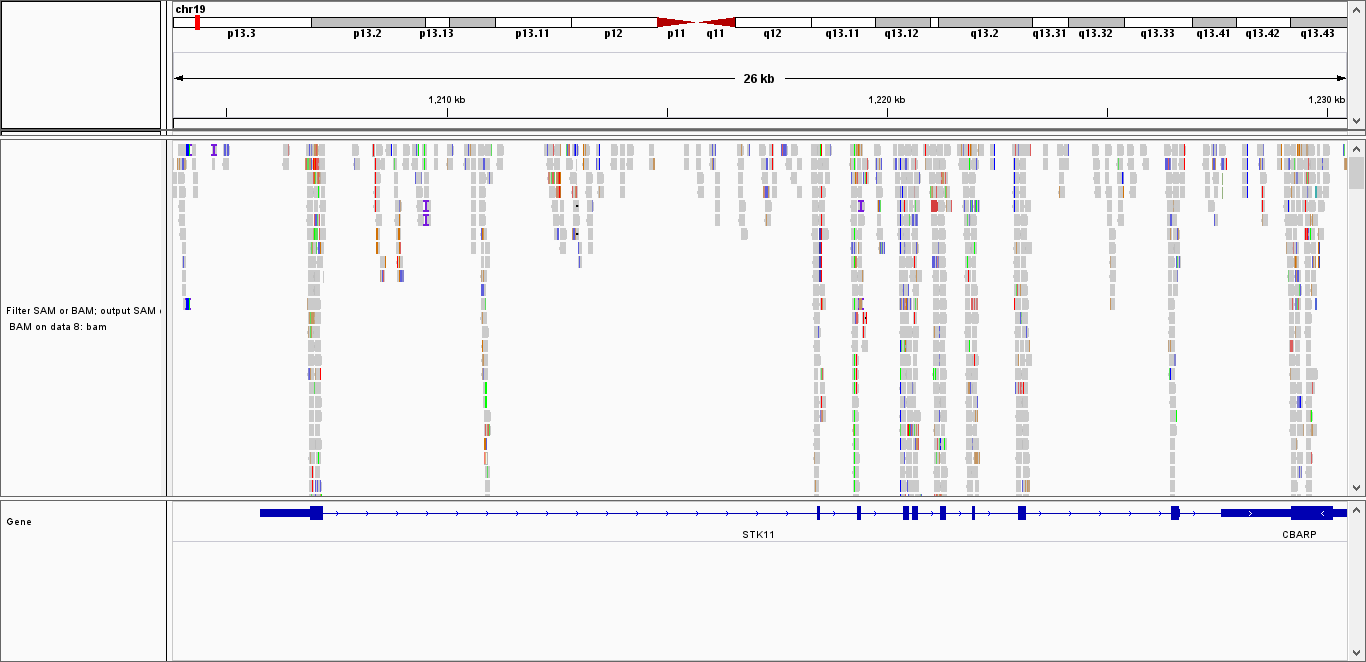
Integration of WXS data with the sequencing reads of the patients in The Cancer Genome Atlas.
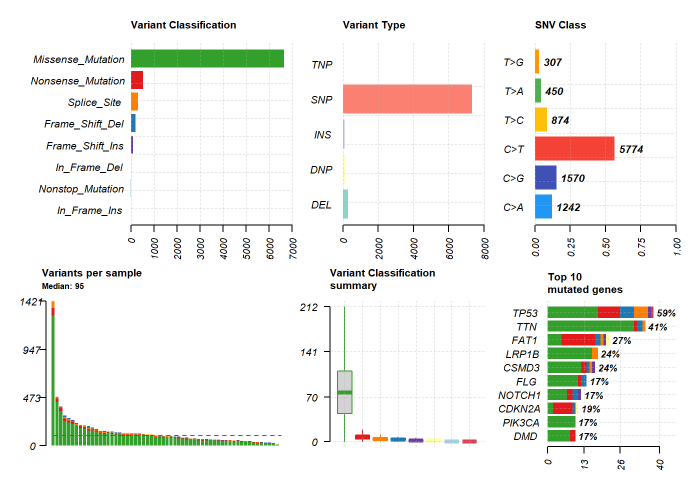
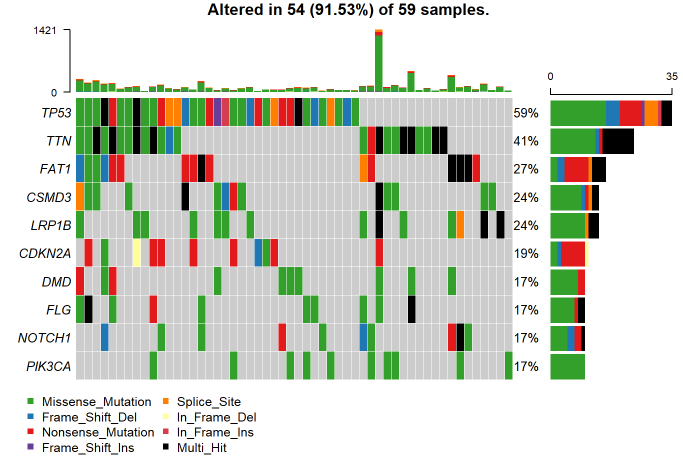
At the end of the session, we took feedback from everyone who attended the session which was all positive and mostly about how Galaxy is a convenient platform for the analysis of genomic data. A group photo of everyone who attended and organized the training session:



