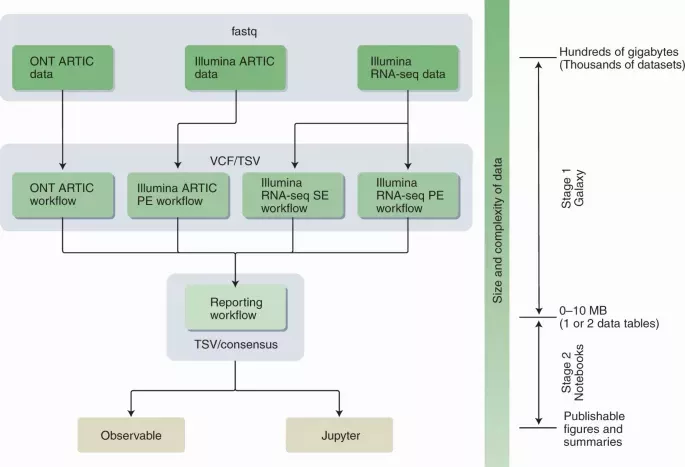
The COVID-19 pandemic is the first health crisis characterized by large amounts of genomic data. Computational infrastructure can be a bottleneck for data analysis, amplifying global inequalities in ability to track SARS-CoV-2 evolution. This is an issue even in developed countries, as computational infrastructure requires expertise in resource procurement, configuration and maintenance. Commercial computational clouds do not fully address the problem because these resources must still be configured and funded. Furthermore, commercial clouds are predominantly US-based and many countries have policies making payments to foreign providers impractical. In developing countries, research computing infrastructure is rare and researchers often cannot afford commercial cloud-based computation. Here, we present the COVID-19 effort by the Galaxy Project, which pools free worldwide public computational infrastructure, making the analysis of deep sequencing data accessible to anyone while also providing an analytical framework for global pathogen genomic surveillance based on raw sequencing-read data.
Read more about our COVID-19 efforts in our summary document and at galaxyproject.org.




